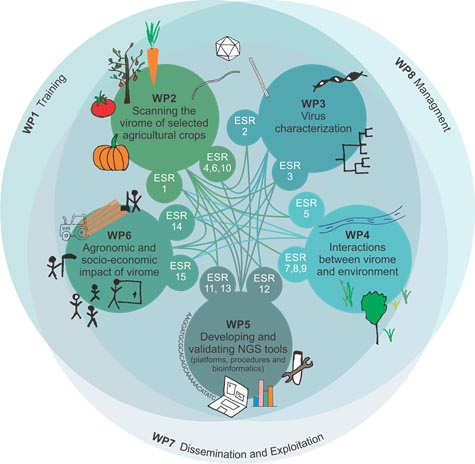
What is INEXTVIR?
INEXTVIR is implemented by a European Consortium of universities, research institutions and companies in Belgium, France, Spain, Slovenia and the UK. It offers fully funded 15 PhD positions in a transdisciplinary network of research and training aimed at accelerating the start of the applicants’ scientific career.
Plant viruses cause 50% of the emerging plant diseases globally and pose an important threat to many agricultural crops. Losses are estimated at €15 to 45 billion per year through lower yields and reduced product quality. That is why Inextvir seeks to generate a better understanding of viral communities and their role in agricultural ecosystems by using the latest advances in high throughput sequencing (HTS) technologies coupled with modern big data analytical approaches and socioeconomic analysis. The project provides a timely opportunity to change our approach to plant health and improve our ability to overcome global agricultural, food security and environmental challenges.