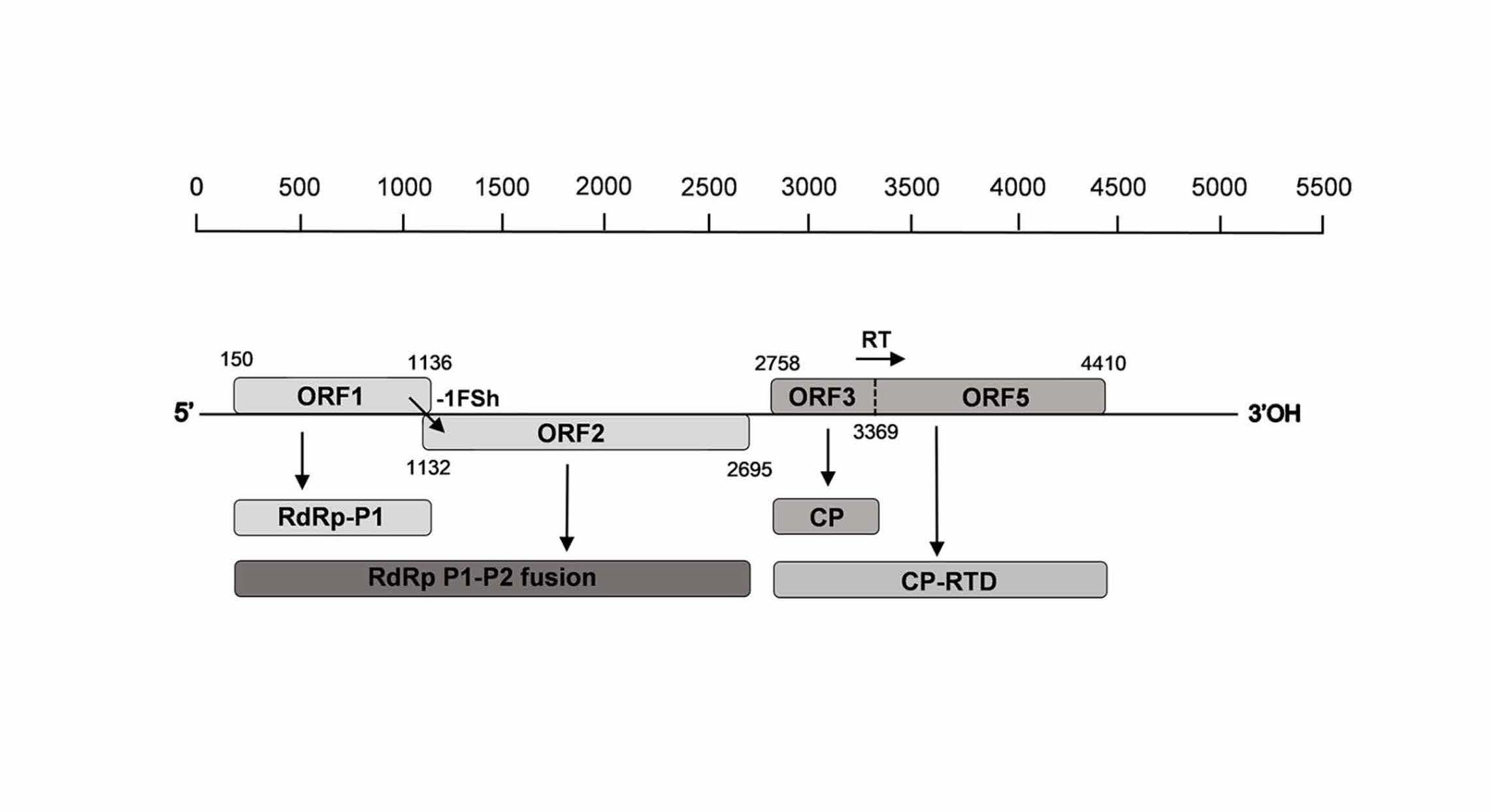
Carrot populations in France and Spain host a complex virome rich in previously uncharacterized viruses
Deborah Schönegger, Armelle Marais, Bisola Mercy Babalola, Chantal Faure, Marie Lefebvre, Laurence Svanella-Dumas, Sára Brázdová, Thierry Candresse. https://doi.org/10.1371/journal.pone.0290108. Using a HTS approach, we analyzed the virome of 51 cultivated and wild carrot populations from France and central Spain. A rich virome comprising 45 viruses of which 25 are novel was identified. Most of the novel viruses showed preferential association with wild carrots, indicating the role of wild carrots as reservoir of viral diversity. The carrot virome proved rich in viruses involved in complex interactions such as poleroviruses, umbraviruses and associated satellites, which can be the basis for further investigations of virus-vector-host relationships
Read more...